ERF Transcription Factors Drive C4 Photosynthesis Evolution, Collaborative Study Reveals
C4 photosynthesis, a highly efficient adaptation that evolved from C3 ancestors over 35 million years ago, has emerged independently ~70 times in flowering plants—a hallmark of convergent evolution. C4 plants outperform C3 species under stress, achieving superior light, nitrogen, and water use efficiency. Engineering this pathway into C3 crops holds transformative potential for global food security, often termed the "Apollo Program" of agricultural innovation. Yet, progress remains limited due to the complexity of C4 evolution.
To address this, Dr. Xin-Guang Zhu’s team at the Chinese Academy of Sciences pioneered an evolution-guided strategy, decoding molecular mechanisms behind C4 innovation. By reconstructing C4’s evolutionary trajectory and leveraging genetic screens, they aim to engineer "C4 prototypes" in crops. The core of this strategy lies in deciphering the molecular evolutionary mechanisms underlying C4 photosynthesis.
Recently, Dr. Xin-Guang Zhu’ team, in collaboration with researchers from the Institute of Biotechnology, Chinese Academy of Agricultural Sciences (led by Dr. Tiegang Lu) and the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (led by Dr. Chengzhi Liang), published a groundbreaking study titled “A dominant role of transcriptional regulation during the evolution of C4 photosynthesis in Flaveria species” in Nature Communications. The study systematically unravels the evolutionary process of genes encoding C4 photosynthetic enzymes in the genus Flaveria and highlights the pivotal role of ERF family transcription factors in driving C4 evolution.

The genus Flaveria, which includes C3 species, C4 species, and intermediate types between the two, serves as an ideal model for studying C4 photosynthesis. The research team constructed chromosome-level high-quality genomes for five representative Flaveria species (one C3 species, three C3-C4 intermediate species, and one C4 species) for the first time, systematically unraveling the evolutionary trajectory of C4-related genes (Fig. 1).
1. Retrotransposon-Driven Genome Expansion and Gene Duplication
Chromosome-level genome assemblies of five Flaveria species revealed a striking 4-fold genome size expansion in C4 lineages (~0.5 Gb in C3 species to ~2 Gb in C4 species), driven primarily by proliferating long terminal repeat retrotransposons (LTR-RTs), which constitute 56.6% of the C4 genome. Despite this expansion, protein-coding genes remained conserved (~35,000 genes), with C4 metabolic genes (e.g., CA, PEPC) retaining ancestral chromosomal positions across species (Fig. 1).
Notably, C4-specific gene duplications—such as the tandem amplification of PEPC1 in the C4 species F. trinervia—were linked to LTR-RT activity. Flanking sequences of new PEPC1 copies showed retrotransposon signatures (inverted repeats, target site duplications), implicating retrotransposition as a key driver of functional innovation.
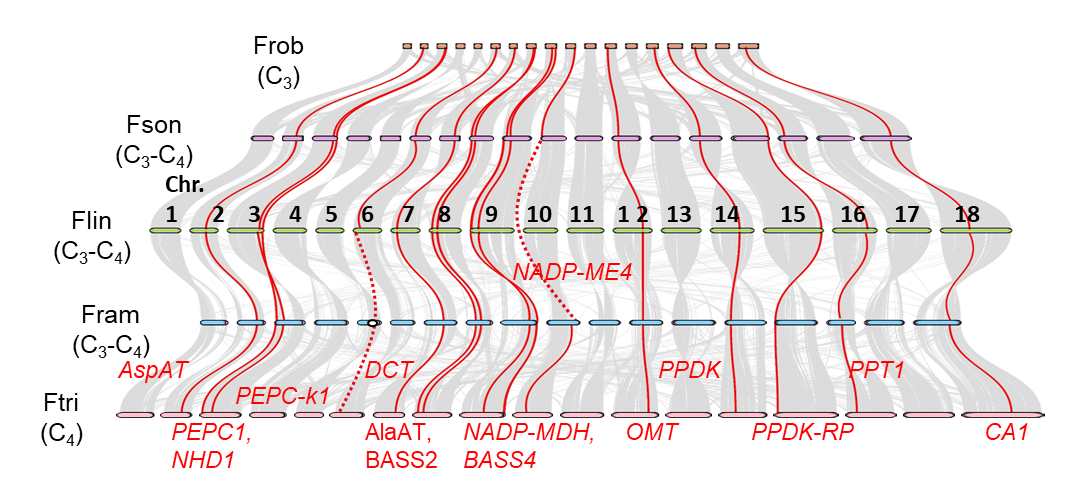
Fig. 1 | Conserved chromosomal locations of C4 genes across Flaveria species
2. Transcriptional Upregulation as the Evolutionary Lever
While C4 metabolic genes exist in ancestral C3 species, their low expression precludes photosynthetic functionality. Comparative analyses demonstrated that transcriptional upregulation, not translational enhancement, propelled C4 evolution. although both transcript levels and protein levels of C4 genes were elevated in C4 species, the magnitude of increase differed. The protein-to-RNA ratio (PTR) of C4 genes decreased significantly in C4 species (Fig. 2). Subsequent translatome analysis confirmed that translation efficiency of C4 genes showed no significant difference between the C3 species F. robusta and the C4 species F. trinervia, further supporting transcriptional regulation as the key driver of C4 gene evolution.
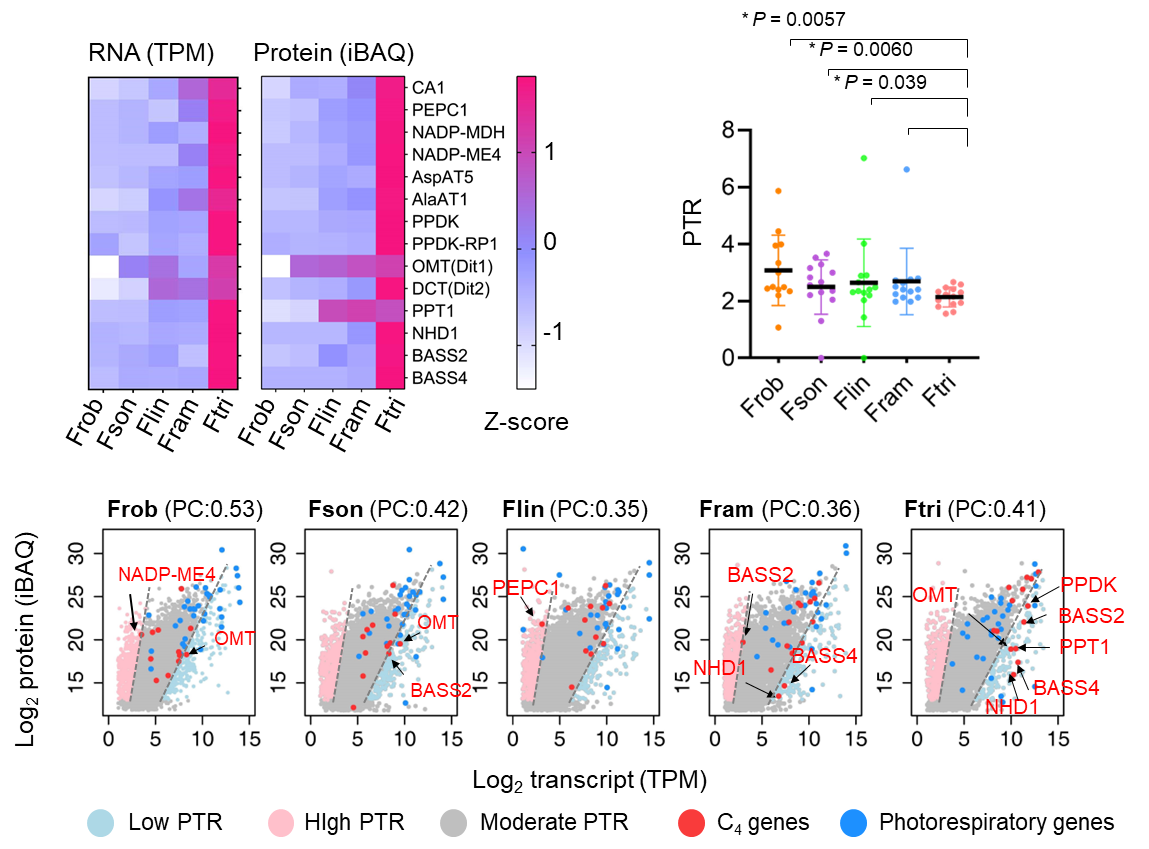
Fig. 2 |Transcriptional upregulation dominates over protein-level increases in C4 gene evolution
3. ERF Transcription Factors: Master Regulators of C4 Adaptation
By comparing C4 gene regulatory networks across species, the team discovered that ethylene response factors (ERFs) —a family of transcription factors typically involved in abiotic stress responses—were preferentially recruited to regulate C4 genes in the C4 species F. trinervia. Consistently, C4 gene promoters in the C4 species acquired abundant ERF-binding cis-regulatory elements, which were located in open chromatin regions (Fig. 3). This repurposing of stress-responsive ERFs aligns with the hypothesis that C4 photosynthesis evolved as an adaptive innovation, allowing ancestral C3 plants to thrive under environmental pressures such as drought, high light, and low atmospheric CO2 conditions, where ERFs’ ancestral role in stress signaling likely primed their recruitment for photosynthetic adaptation.
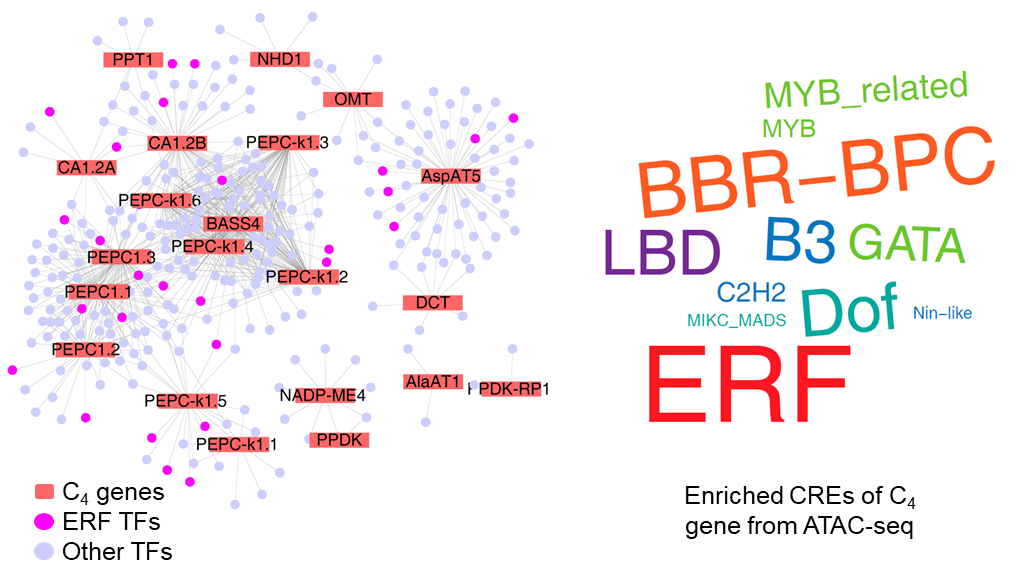
Fig.3| Expanded recruitment of ERF transcription factors in regulating C4 genes in Flaveria C4 species
The team has established a Flaveria genomic data portal (accessible at https://db.cngb.org/codeplot/datasets/flaveria) on China’s National GeneBank platform, offering open access to datasets and accelerating progress in C4 photosynthesis research.
Link:https://www.nature.com/articles/s41467-025-56901-y
Research Background: A Decade of Pioneering Work on C4 Photosynthesis Evolution
Dr. Xin-Guang Zhu’s team has long been dedicated to advancing plant photosynthetic efficiency research, focusing on photosynthetic systems biology, high-efficiency gene mining in crops, and C4 photosynthesis evolution and prototype engineering. Over the past decade, their systematic studies on Flaveria—a model genus for C4 evolution—have yielded a series of new insights:
1. Constructed the phylogenetic tree of representative Flaveria species (Lyu et al., 2015, BMC Evolutionary Biology).
2. Proposed that high-expression gene copies are prerequisites for C4 pathway recruitment (Lyu et al., 2020, Frontiers in Plant Science).
3. Identified two critical evolutionary transition points during C4 evolution (Lyu et al., 2021, Scientific Reports).
4. Revealed that some C3-C4 intermediates represent parallel evolutionary states rather than transitional steps toward C4 (Lyu et al., 2023, Plant Communications).
5. Demonstrated that reduced Stomagene expression drives stomatal density decline in C4 species (Zhao et al., 2022, Plant Physiology).
6. Established that elevated α-ketoglutarate (α-KG) levels are essential for C4 pathway establishment (Tang et al., 2024, Plant Physiology).
7. Uncovered the central role of ERF transcription factors in C4 evolution.
Collectively, these studies have reconstructed the molecular evolutionary trajectory of C4 photosynthesis, delineated key evolutionary stages and engineering targets, and established a theoretical framework for evolution-guided C4 prototype design in crops.