Pangenome Study Unveils Wild and Cultivated Rice's Genetic Diversity
A landmark genomic study led by Professor Bin Han and his team at the Center for Excellence in Molecular Plant Sciences of the Chinese Academy of Sciences, has decoded the genetic architecture and diversity of rice, offering new insights into its evolutionary and domestication history. Published online in Nature on April 16 under the title "A pangenome reference of wild and cultivated rice", the research constructed an unprecedented pangenome map of wild and cultivated rice, providing a powerful resource for breeding and agricultural innovation.
Facing both climate change and a rapidly growing global human population, Asian cultivated rice (Oryza sativa), a staple food for billions worldwide, urgently needs to sustainably boost its resistance and yield. While domesticated rice has been extensively studied, its wild progenitor, Oryza rufipogon—shaped by thousands of years adaptation to diverse environments—has remained largely underexplored, leaving critical gaps in understanding the full genetic potential of wild rice.
To address this knowledge gap, the research team obtained 145 geographically and genetically diverse rice genomes, including 129 wild accessions and 16 cultivated varieties. Mainly using advanced PacBio high-fidelity (HiFi) sequencing technology and computational methods, they created the highest resolution “pangenome” to date that, capturing the full genetic landscape of wild rice and revealing hidden variations critical for crop improvement.
The study uncovered 3.87 billion base pairs of novel genetic sequences absent from the single acknowledged reference genome (O. sativa ssp. japonica cv. Nipponbare), along with 69,531 genes collectively spanning the pangenome. Remarkably, nearly 20% of these genes exist only in wild rice, many linked to disease resistance and environmental adaptation. These genes represent a ‘genetic goldmine’ that could help develop modern rice varieties capable of withstanding pests, diseases, and the challenges of a changing climate.
Using high-quality genome sequences, this study conducted haplotype analysis of early key domestication genes in various groups of Asian cultivated rice, and proved that all domestication loci were derived from the japonica ancestor Or-IIIa. The findings strongly support the hypothesis that all Asian cultivated rice groups underwent a single initial domestication event, providing crucial evidence for a long-standing scientific debate. In addition, the research team identified extensive gene flow among cultivated rice groups in South Asia, leading to the classification of a newly identified subpopulation, intro-indica, and successfully mapping a comprehensive roadmap of rice evolution and domestication.
The study also explored the genetic divergence between indica and japonica, the two main subspecies. Researchers identified over 850,000 single-nucleotide polymorphisms and 13,000 presence–absence variations between them. Their findings suggest that these variations mainly originated from the divergence of their respective ancestors and the existence of a larger genetic bottleneck in japonica. This understanding opens new opportunities to combine beneficial genes from different rice subspecies.
The near-saturated pangenome dataset, integrating valuable wild genetic resources, provides a powerful foundation for agricultural researchers and plant breeders. Scientists can mine favorable alleles, trace the origin of key genes and enhance understanding of rice environmental adaptation and phenotypic plasticity. Moreover, this study offers a roadmap to develop rice varieties that can withstand climate extremes, require fewer resources, and produce higher yields. In a companion Research briefing titled “Unlocking the diversity of wild and domesticated rice”, Nature’s editors commended the study for its importance to future food security.
This research was funded by the National Natural Science Foundation of China, the Strategic Priority Research Program of the Chinese Academy of Sciences and the Ministry of Agriculture and Rural Affairs.
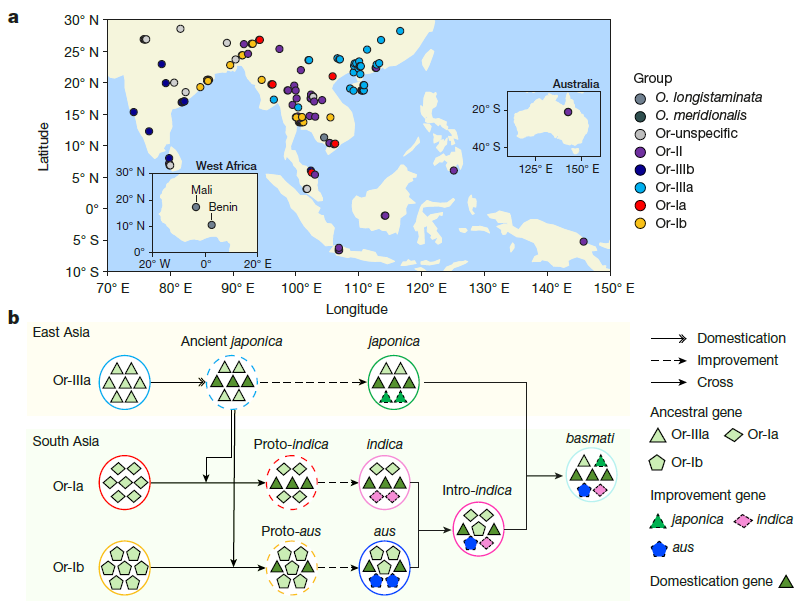
Pangenome and evolutionary analysis of Asian cultivated and wild rice. a, Geographical distribution of 133 wild-rice genome sequences, with dots coloured by group. ‘Or’ groups are genetically related groups, or clades, of Oryza rufipogon; Oryza longistaminata and Oryza meridionalis are related species. The inset maps show enlarged views of the West Africa and Australia regions. b, Evolutionary history of Asian cultivated rice. Milestones in the domestication and improvement of various groups of Asian cultivated rice are shown, highlighting notable cross-breeding events.
Article Link: https://doi.org/10.1038/s41586-025-08883-6
Contact:
Dr. Bin Han, Principal Investigator
Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences
Email: bhan@cemps.ac.cn