Chloroplast ROS: the smoking gun to decipher plant-specific stress/cell death pathways
In 2004, Prof. Klaus Apel (1942-2017) and his research team at ETH Zurich reported an inspiring story, the genetic basis of chloroplast-mediated programmed cell death controlled by the chloroplast-driven singlet oxygen (1O2) and its sensor protein EXECUTER1 (EX1) (Wagner et al., 2004 science). The failure of 1O2 sensing through either the modification or deletion of the sensor domain of EX1 abolishes the expression of nuclear 1O2-responsive genes and cell death (Wang et al., 2016 PNAS; Dogra et al., 2019 Nat Commun), suggesting that chloroplasts are involved in a genetically controlled cell death via retrograde signaling. In 2012, they also identified a peculiar phenotype in well established Arabidopsis mutants defective in chloroplast binary fission (Simkova et al., 2012 Plant Journal). These mutants, carrying gigantic chloroplasts (GC), such as crl, arc6, ftsz1-1, and pdv2, appear to develop a light-dependent localized foliar cell death in photosynthetic tissue. This phenotype mirrors that of typical lesion mimicking mutants (LMMs), which have widely been used as bio-tools to unravel molecular mechanisms underlying cell death and immune responses in land plants.
A team of scientists from the Shanghai Center for Plant Stress Biology, Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, led by Prof. Chanhong Kim, sheds new light on the underlying mechanism of GC-mediated cell death response through a combination of reverse and forward genetic approaches.
Remarkably, the reverse genetic approach excludes any potential role of stress hormones (e.g., SA and JA) in triggering cell death in GC mutants. Instead, transcriptome and in silico analyses suggest that reactive electrophile species (RES) generated via oxidation of unsaturated fatty acids or lipid peroxidation-driven signaling may prime cell death. Consistently, the crl-suppressor spcrl4 contains a causative mutation in the nuclear gene encoding the chloroplast-localized FATTY ACID DESATURASE 5 (FAD5) catalyzing the conversion of palmitic acid (16:0) to palmitoleic acid (16:1) (Figure 1). The loss of FAD5 in the crl mutant attenuates the levels of RES and/or lipid peroxidation due to the reduced levels of the palmitic acid-driven unsaturated fatty acids, prime targets of reactive oxygen species. The fact that fad5 also compromises the expression of immune-related genes and cell death in other GC mutants substantiates the presence of an intrinsic retrograde signaling pathway, priming the autoimmune responses in a FAD5-dependent manner.
This work, entitled "FATTY ACID DESATURASE 5 is Required to Induce Autoimmune Responses in Gigantic Chloroplast Mutants of Arabidopsis" is now published by The Plant Cell (10.1105/tpc.20.00016). The study is supported by the Strategic Priority Research Program from the Chinese Academy of Sciences (Grant No. XDB27040102) and the National Natural Science Foundation of China (NSFC) (Grant No. 31871397) to C.K.
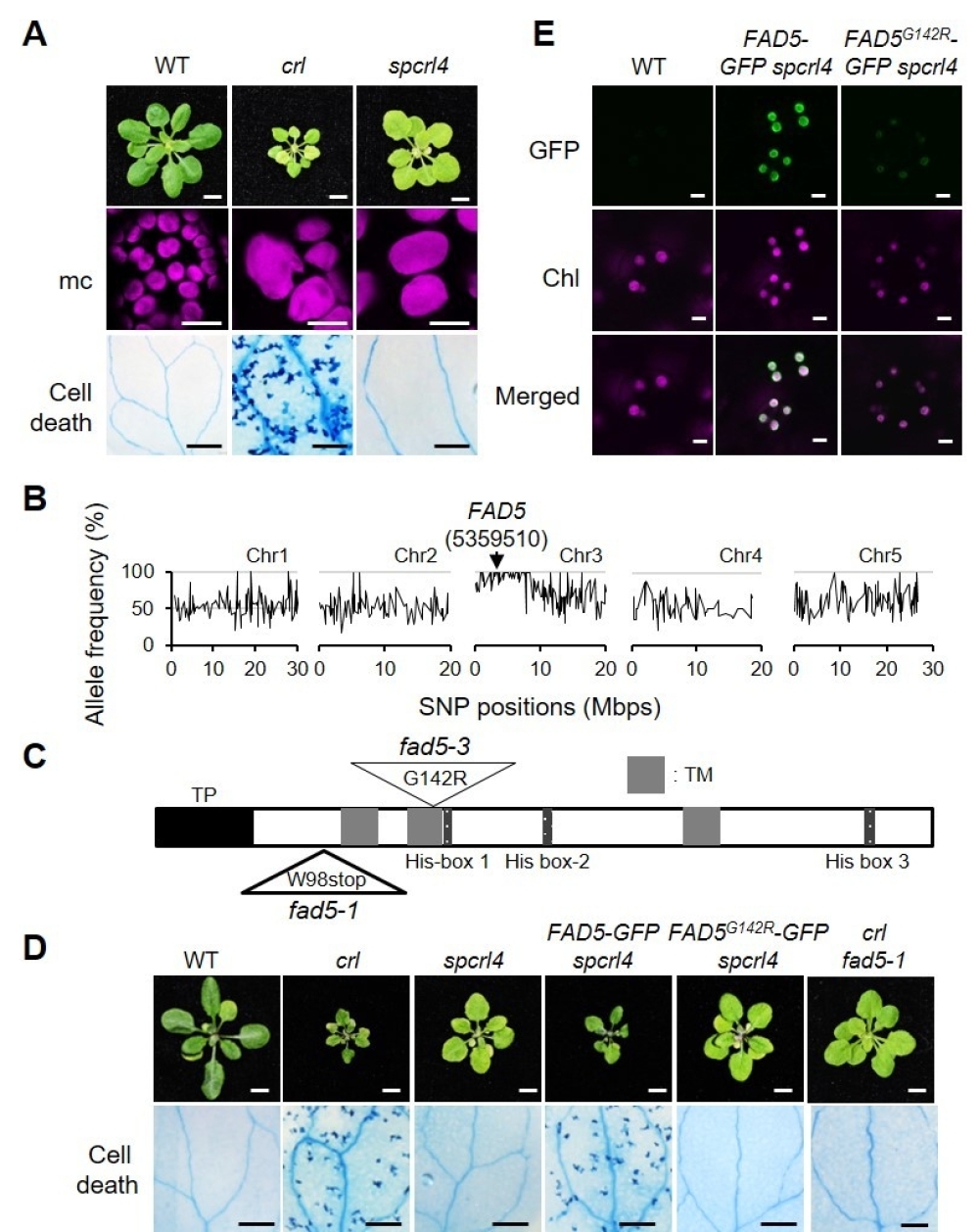
Figure. Inactivation of FAD5 Abrogates Cell Death in crl.
(A) Representative images of 21-d-old plants grown on soil under CL (Continuous Light) are shown in the top row (Bars = 0.5 cm). Confocal images of chlorophyll fluorescence in mesophyll cells (mc) in 5-d-old seedlings are shown in the middle row (Bars = 15 μm). Cell death in cotyledons (10-d-old seedlings) was visualized by TB staining and representative results are shown in the bottom row (Bars = 1 mm). (B) Whole-genome sequencing of spcrl4 in comparison with crl identified a mutation in FAD5 at position 5359510 in chromosome 3. Mbps, million base pairs. (C) A schematic illustration of FAD5 shows a transit peptide (TP, black), three transmembrane domains (TM, grey), and three His-box domains (dark grey). The mutation positions for fad5-1 and fad5-3 are indicated with triangles (W, Trp; stop, stop codon; G, Gly; R, Arg). (D) Complementation assay. Images are representative of 21-d-old plants grown on soil under CL and shown at the same scale (Bars in top row = 0.5 cm). Cell death was visualized by TB staining in the cotyledons of 10-d-old seedlings grown under CL, and a representative image from each genotype is shown (Bars in bottom row = 1 mm). (E) Subcellular localization of FAD5- GFP and FAD5G142R in 5-d-old seedlings of stable spcrl4 transgenic lines grown on soil under CL (Bars = 5 μm).
Article link: http://www.plantcell.org/content/early/2020/08/13/tpc.20.00016
Contact:
Shengwei Li, PIO of PSC
Email: swli@psc.ac.cn
Shanghai Center for Plant Stress Biology, Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences