A novel paradigm of bacterial transcription activation
Textbooks categorically describe allosteric transcriptional activation mechanisms in terms of the ligand-induced conformational changes that enable the transcription factor to contact the polymerase essentially recruiting polymerase or accessory factors into a transcriptionally active complex. There is much support for these types of mechanisms, so much so that protein-polymerase contacts are often described as a universal mechanism for allosteric transcriptional control.
A research work published at Nature Chemical Biology today (https://www.nature.com/articles/s41589-020-00653-x) unequivocally shows for the first time that a transcription factor can turn on transcription in the absence of any contacts with the polymerase. By solving the cryo-EM structure of the E. coli transcription activation complex CueR-TAC that comprises CueR, a MerR-family transcription factor, promoter DNA, and RNAP, the authors present clear evidence that, unlike most transcription factors, CueR has no interactions with RNAP. The structure shows that CueR works in concert with RNAP to remodel the stereochemistry of the entire promoter via formation of four significant kinks in DNA structure. This switch from B-DNA to highly kinked DNA dramatically changes the overall stereochemistry of the promoter, putting the -10 element (a key promoter element essential for transcription initiation) adjacent to the active site residues responsible for formation of the transcription bubble, and thus, turns on transcription.
The results provide the strongest case yet for transcription control through the modulation of DNA stereochemistry and topology.
This work was collaborated by three research groups, Dr. Yu Zhang’s lab (at CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology & Ecology, Chinese Academy of Sciences), Dr. Yu Feng’s lab (at Zhejiang University school of Medicine), and Dr. Thomas O’ Halloran’s lab (at the Chemistry of Life Processes Institute, Northwestern University). This work was supported by the grants from National Natural Science Foundation of China, Strategic Priority Research Program of the Chinese Academy of Sciences, and the National Institutes of Health of the United States.
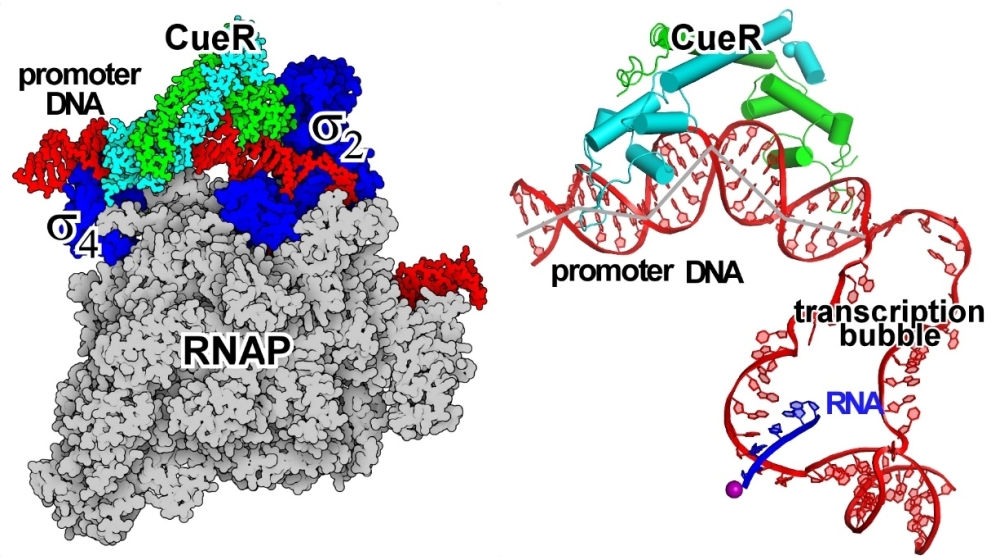
Figure 1. The cryo-EM structure of E. coli CueR-TAC. (Left) CueR locates at the upper face of promoter DNA, while RNAP locates at the lower face of promoter DNA.
(Right) CueR and RNAP induce four kinks at the upstream double-stranded promoter DNA.
Link: https://www.nature.com/articles/s41589-020-00653-x
Contact:
Dr. Yu Zhang, Professor
Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences/Shanghai Institute of Plant Physiology and Ecology (SIPPE), CAS
Email: yzhang@sippe.ac.cn