A novel mechanism coupling H3K27me3 recognition with transcriptional repression through BAH-PHD-CPL2 complex
Scientists at the Shanghai Center for Plant Stress Biology, Center for Excellence in Molecular Plant Sciences of the Chinese Academy of Sciences, have discovered that a novel histone reader complex which can recognize H3K27me3 and repress transcription through dephosphorylation of Pol II. This study was published on 4th December, 2020 in Nature Communications.
H3K27me3 is an important histone post-translational modification (PTM) involving in the most biological processes. In traditional views, Polycomb Repressive Complex 2 (PRC2) was responsible for the deposition of H3K27me3 and PRC1 can bind H3K27me3, catalyzing H2Aub and further promoting chromatin condensation to block the accessibility of transcriptional complex. However, this model had been challenged by several recent reports in which many new H3K27me3 reader components were found.
Yizhe Zhang, Jianlong Yuan and colleagues in Drs. Cheng-Guo Duan’s, Jiamu Du’s and Jian-Kang Zhu’s labs identified a new protein complex containing BAH protein AIPP3, PHD protein AIPP2 or PAIPP2, and Pol II phosphatase CPL2 (BAH-PHD-CPL2 complex, BPC complex). The bpc mutants showed the photoperiod-independently early flowering time and developmental defect, indicating the indispensable role of BPC complex in plant development and flowering regulation. Genetic assays suggested that BPC regulates flowering through constitutively repression of FT expression.
Through structural and biochemical assays, they found that the AIPP3-BAH and the AIPP2/PAIPP2-PHD cooperate to read H3K27me3 and unmodified H3K4 histone marks, respectively, in Arabidopsis. ChIP-seq and mRNA-seq data showed that BPC complex directly binds and silences a substantial subset of H3K27me3-enriched loci, including a number of development and stress response-related genes. Interestingly, H3K27me3 deposition was not obviously changed between Col-0 and bpc mutants at selected target genes, indicating that BPC complex acts in downstream pathway of H3K27me3 deposition.
CPL2 has been reported as a plant-specific phosphatase to eliminate phosphorylation on the Ser5 (ser5P) of Pol II carboxyl terminal domain (CTD). Through a published NET-seq data of Pol II, they found that only a sharp peak of unphosphorylated Pol II signals was observed at the TSS region of the selected target genes but phosphorylated Pol II not, indicating BPC complex interrupts Pol II initiation rather than recruitment. They observed higher accumulations of phosphorylated Pol II in bpc mutants. More importantly, they found BAH-PHD module is indispensable for chromatin localization of CPL2.
In summary, this study first connected the recognition of H3K27me3 with Pol II dephosphorylation, suggesting a unique H3K27me3-mediated epigenetic silencing mechanism independent with canonical PRC2-PRC1-H2Aub pathway. Although CPL2 is a plant-specific phosphatase, the crosstalk between histone modification and Pol II CTD codes may exist widely in eukaryotic system. Recently years, BAH-H3K27me3 module was discovered in plants, fungi and animals. However, their function showed different. In plant, the other two BAH proteins EBS and SHL bind H3K27me3 region and participate in PRC1-H2Aub pathway, redundantly with LHP1. On the other side, mammalian protein BAHD1 and BAHCC1 (BAHD2) also can recognize H3K27me3 through their BAH domain. Interestingly, BAHD1 interacts with PRC2 complex and responsible for H3K27me3 deposition while BAHCC1 associates with HDAC1, maintaining a hypoacetylated chromatin state at H3K27me3-enriched genes. Above studies indicated the fact that the H3K27me3-mediated silencing pathway is much more complicated than expected.
Funding: This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27040203) and by the National Natural Science Foundation of China (31570155).
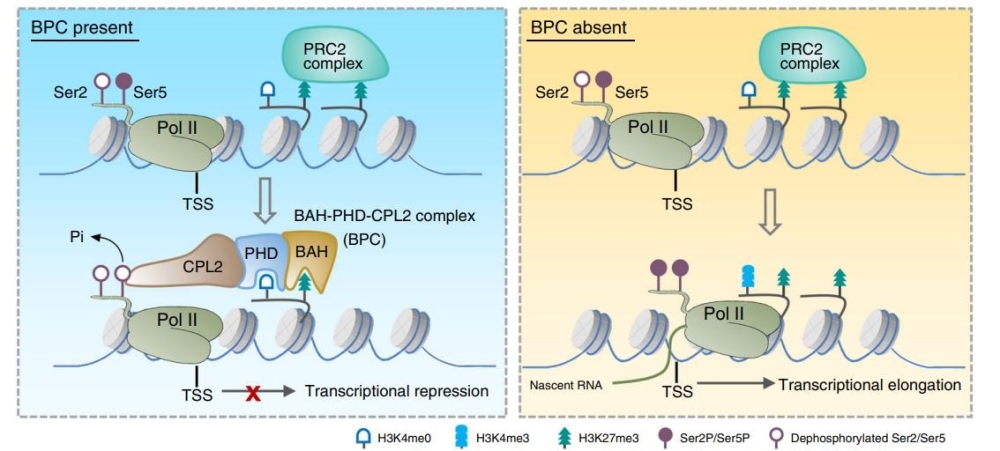
Figure: A working model of the BPC complex-mediated transcription repression
Link: https://www.nature.com/articles/s41467-020-20089-0