Winning Gene Combination Takes All
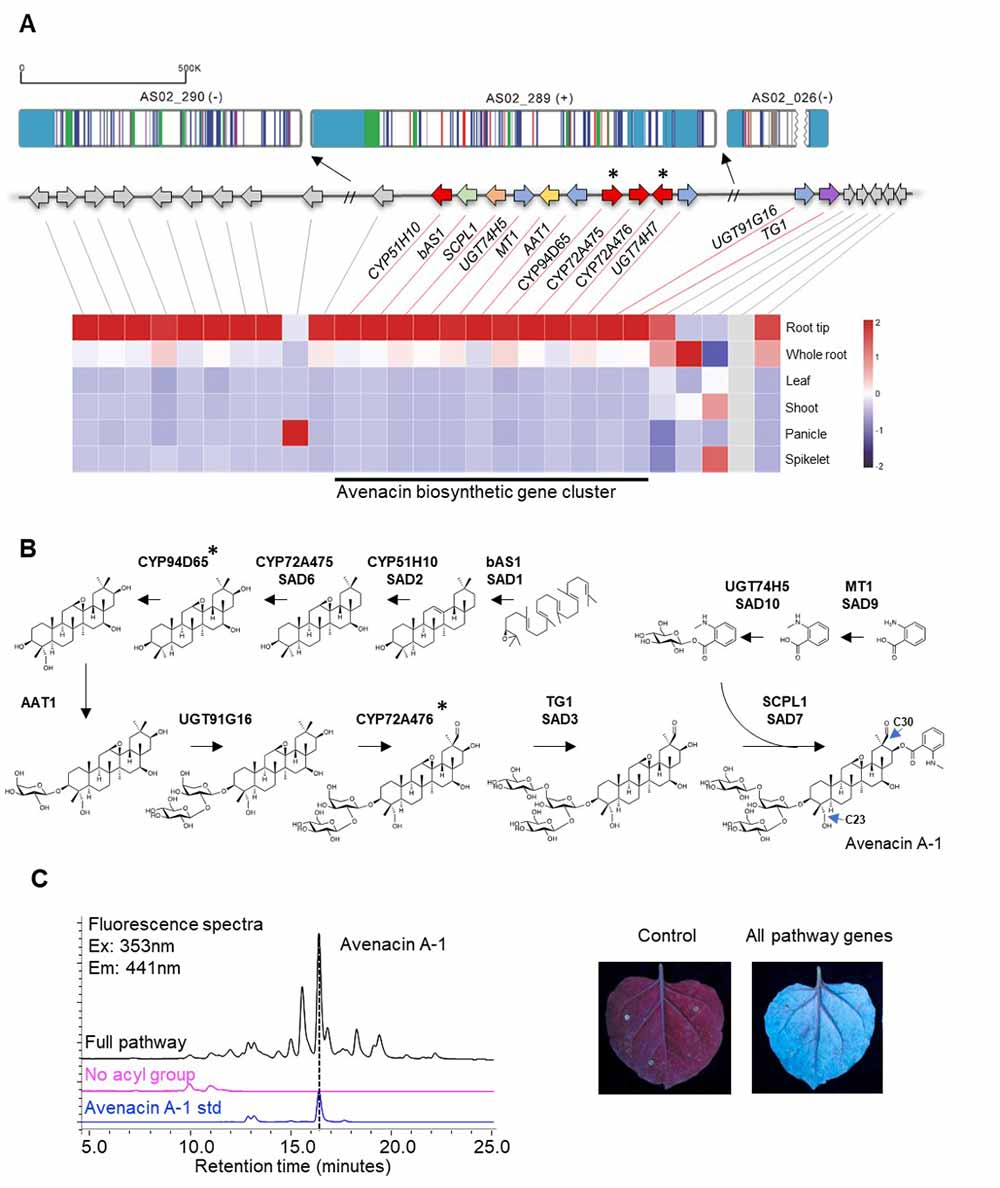
Avenacins are antimicrobial compounds synthesised in the roots of oats where they offer protection against soil-borne diseases such as take-all. This fungal pathogen causes huge yield losses in wheat and there is no effective means of control. Wheat and other cereals and grasses do not make these compounds but a better understanding of how they are produced in oat will give crop scientists knowledge they need to create disease resistant lines of wheat using modern technologies. Earlier experiments had characterised and cloned ten avenacin biosynthetic pathway genes found in the oat genome.
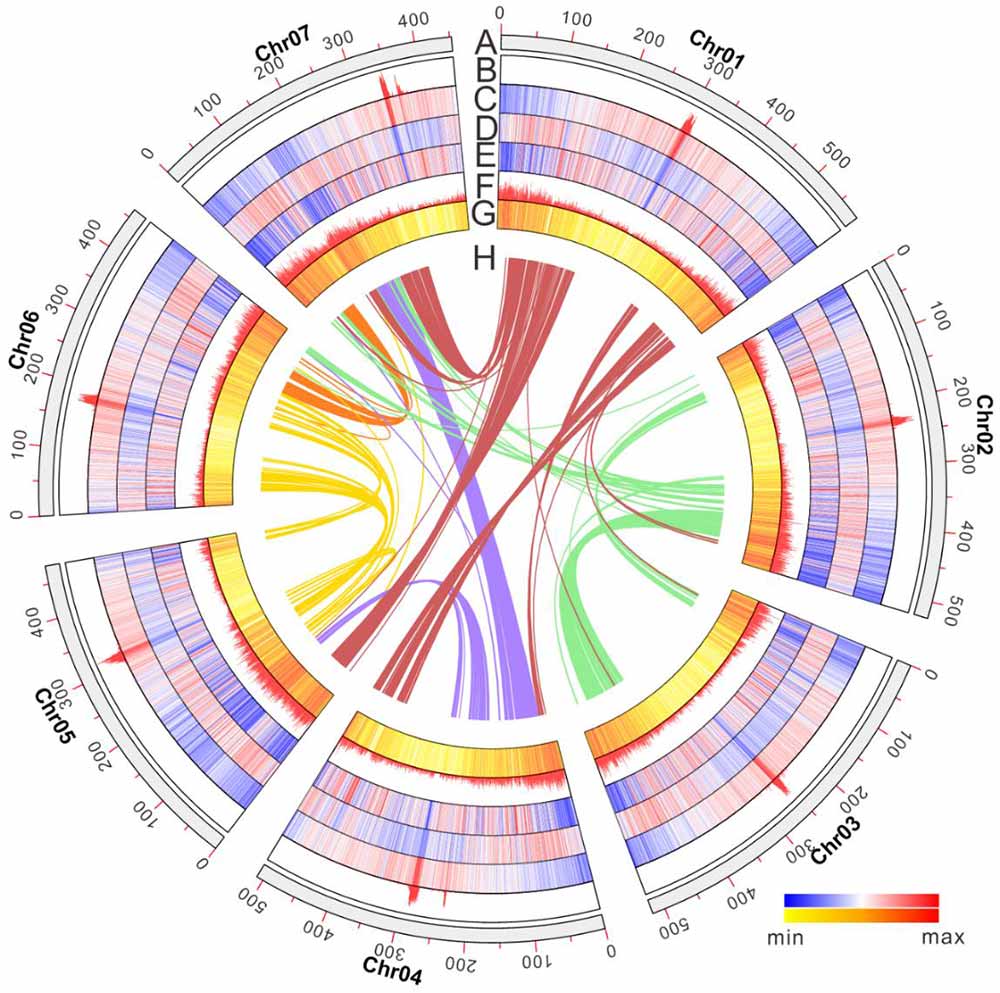
On May 7st, 2021, the study ‘Subtelomeric assembly of a multi-gene pathway for antimicrobial defense compounds in cereals’ supported by CEPAMS – a collaboration between the John Innes Centre and the Chinese Academy of Sciences appears in Nature Communications. This research elucidated the complete pathway, encoded by 12 genes using a genomics-driven approach, with sequencing carried out by Professor Bin Han’s group at the Chinese Academy of Sciences. They found that genes are clustered next to each other in the genome like beads on a string and organised along the chromosome approximately in the same order as the biosynthetic pathway – like a recipe written out in order of ingredients. The avenacin gene cluster is located very close to the end of one arm of chromosome 1 of oat. It is arranged such that the early pathway genes are closer to the end of the chromosome (the telomere) and the late pathway genes are further in. The team speculate this may be because gene mutations in the late avenacin pathway can result in the accumulation of compounds that negatively affect plant growth while mutations in the early pathway genes do not. The orientation of these late pathway genes away from the telomere region means the plant is less likely be affected by toxins. Comparison with the sequenced genomes of other cereals and grasses revealed that the avenacin cluster has formed since the divergence of oats from these other plant species which, the researchers presume, is due to a particular set of selective pressures.
Professor Anne Osbourn, joint corresponding author of the research along with Professor Bin Han said: “Our investigations show that plant genomes are able to shuffle and evolve their genes to enable them to adapt to particular stresses – in this case to soil-borne fungal diseases such as take-all. During this process, winning combinations of genes that provide a selective advantage can be recruited and relocated from around the genome and assembled into a cluster like beads on a string. This clustering will enable the winning gene-set to be passed on from generation to generation and mitigate against incomplete inheritance of the pathway genes with associated deleterious effects.”
The study offers the latest example of plant biosynthetic gene clusters for different types of compounds including drugs. Investigations of how widespread these types of genomic organisations are in the Plant Kingdom hinges on the generation of new genome sequences for a wider variety of plants. This should be made possible with the onset of new large-scale genome sequence initiatives such as the Earth BioGenome and Darwin Tree of Life Projects.
Fig1. Avena strigosa genome features.
Fig2. The complete 12-gene avenacin biosynthetic cluster and full pathway reconstitution by transient expression in Nicotiana benthamiana.
Article link: https://www.nature.com/articles/s41467-021-22920-8
Contact:
Bin Han
National Centre for Gene Research, CAS-JIC Centre of Excellence for Plant and Microbial Science (CEPAMS), Centre of Excellence for Molecular Plant Sciences, Chinese Academy of Sciences (CAS), Shanghai 200032, China
Tel: 86-21-54971325
Email: bhan@ncgr.ac.cn