CAS Scientists Unveil a New Paradigm for Transcription by a Two–RNA Polymerase Complex
Teams at the CAS Center for Excellence in Molecular Plant Sciences (CEMPS) led by Yu Zhang and Jiawei Wang, collaborated with Yu Feng at Zhejiang University, have determined the three-dimensional structures of a two-polymerase complex that produces double-stranded RNA.
In a paper published in Science today, the researches show how Pol IV and RDR2 are assembled into a tight two–RNA–polymerase complex and efficiently produce double-stranded RNA using genomic double-stranded DNA as the template.
Eukaryotic organisms encode five multiple–subunit DNA–dependent RNA polymerases. Polymerase (Pol) I, Pol II, and Pol III are conserved in all eukaryotic organisms and mainly responsible for transcription of rRNA, mRNA, and tRNA, respectively. However, Pol IV and Pol V are specific to land plants and are responsible for producing RNAs essential for repressing transposable elements, establishing genome imprinting, and maintaining genome integrity.
The catalytic activity, general transcription factors, and genomic targeting loci of Pol IV are radically different from the rest of polymerases. More importantly, Pol IV together RDR2, directly produces double stranded RNAs templated by the double stranded DNA at the genomic regions containing transposon elements. Determining the structure of Pol IV and RDR2 has been a goal of researchers in the transcription and epigenetic field since the discovery of the two polymerases.
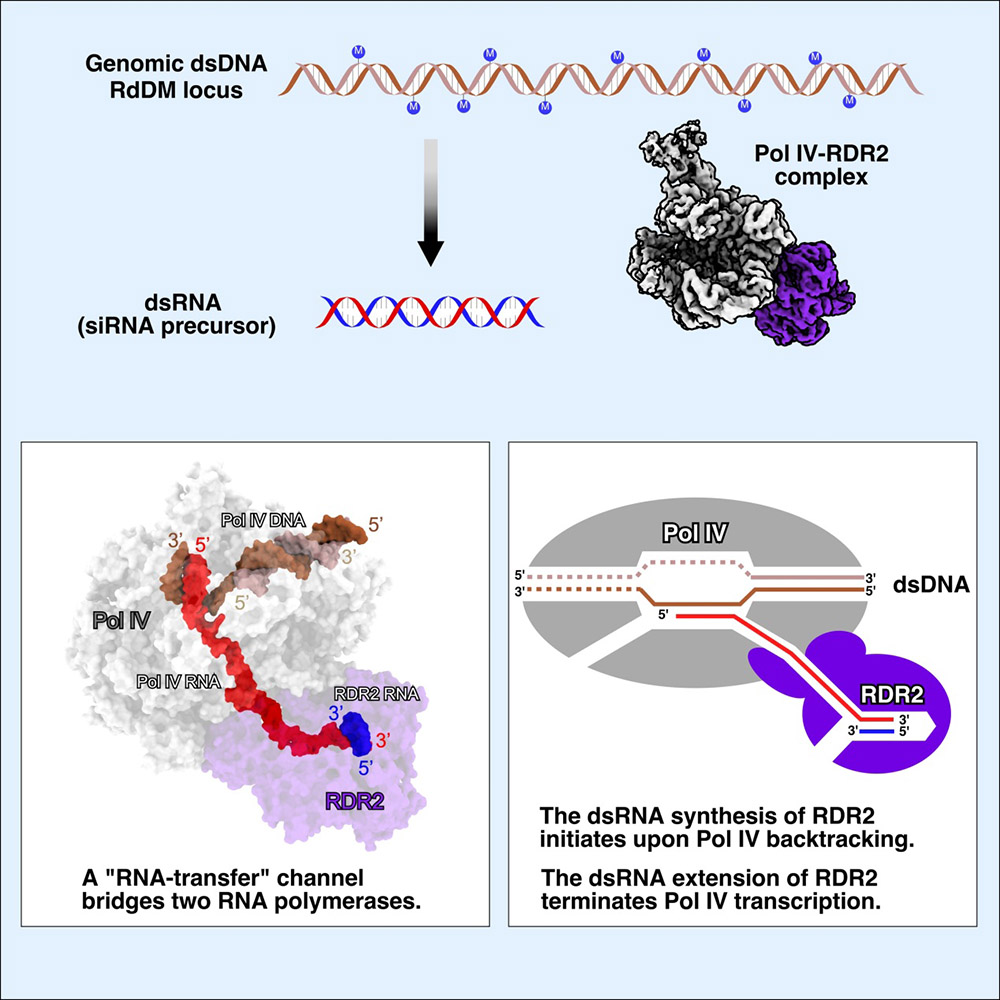
The scientists at CEMPS developed a new method to obtain the endogenous, low-abundance, multiple-subunit Pol IV–RDR2 complex in plant suspension cell cultures and determined the cryo–EM structure of Pol IV–RDR2 holoenzyme and the backtracked transcription elongation complex. The structures reveal that Pol IV and RDR2 form a complex with their active sites connected by an interpolymerase channel, through which the Pol IV–generated transcript is handed over to the RDR2 active site after being backtracked, where it is used as the template for double-stranded RNA (dsRNA) synthesis.
The structures describe a 'backtracking-triggered RNA channeling' mechanism underlying dsRNA synthesis by Pol IV and RDR2. The direct RNA transfer synchronizes transcription cycle of two polymerases allowing efficient dsRNA production. The direct RNA transfer also prevents single–stranded P4–RNA from degrading by multiple cellular RNA nucleases and ensures the substrate specificity of RDR2. More importantly, Pol IV returns to the position where the Pol IV RNA synthesis initiates after completing a transcription cycle, and presumably could initiate another round of Pol IV RNA synthesis to amplify small RNA signals for local or long–distance action.
Prof. Yu Zhang in the Science paper also proposed a previously undescribed means of transcription termination by Pol IV, a dsRNA–induced termination. This termination mode is distinct from all previous transcription termination models of Pol I, Pol II, and Pol III. In this unique model, RDR2 pulls and releases Pol IV RNA in a 3′ to 5′ direction from the Pol IV secondary channel, in sharp contrast to all previous models, in which RNA is released in a 5′ to 3′ direction from the RNA exit channel of RNA polymerases.
The work presents the structure of the fourth member of eukaryotic multiple–subunit RNA polymerases, the first example of 'two–polymerase' complex in all organisms, a novel interpolymerase RNA transfer mechanism, and a new transcription reaction (outputting dsRNA using genomic dsDNA as input).
The cryo–EM structure and working model of the Pol IV–RDR2 complex
Article link:
Contact:
Prof. Yu Zhang, Group leader
Key Laboratory of Synthetic Biology, Center for Excellence in Molecular Plant Sciences (CEMPS), Chinese Academic of Sciences
Email: yzhang@cemps.ac.cn